The BRAINMODEL consortium
VU University and Amsterdam UMC: Functional genomics department
At the Vrije Universiteit and the Amsterdam UMC-Location VUmc the teams of Matthijs Verhage, Ruud Toonen, and Niels Cornelisse will investigate cellular parameters that control the excitation/inhibition (E/I) balance in IPSC derived neurons from patients. They will use patch-clamp electrophysiology and advanced optogenetic tools to measure the amount and strength of contacts (synapses) between excitatory and inhibitory neurons in small networks. The same assays will be used to study the effect of E/I targeting drugs on these parameters and neuronal activity in these networks. Findings at the cellular level will be compared with results from the MEA recordings in larger networks by the Nael Kasri lab and EEG measurements in the same patients by the Hilgo Bruining and Klaus Linkenkaer-Hansen group.
Erasmus MC: Department of Medical Ethics, Philosophy and History of Medicine
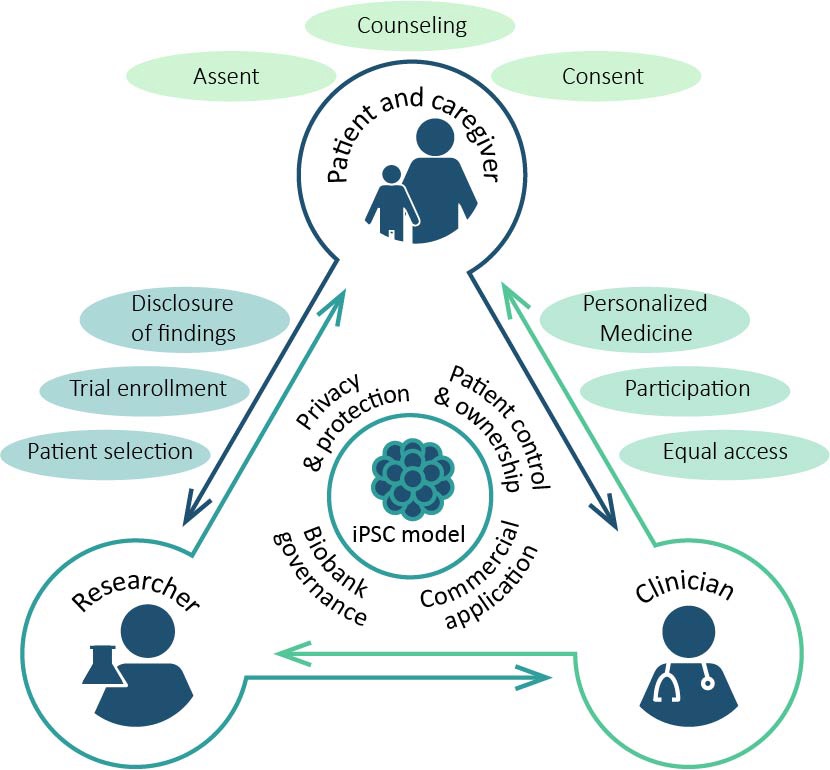
With a research team based at the Department of Medical Ethics, Philosophy and History of Medicine at Erasmus MC, we will investigate the ethical issues associated with BRAINMODEL. From bench to bedside: from the development of neuronal induced pluripotent stem cell (iPSC)-based models in the laboratory to the design of n-of-1 clinical trials and to personalized medicine for patients with monogenic neurodevelopmental disorders (mNDDs). Using qualitative methodologies, we will study the perspectives of patients and clinicians on the use of patient-derived iPSC-models in mNDD care. Also, we will provide practical-ethical guidance within the project, and work with the user/end-user committee to monitor, prioritize and address emerging ethical issues.
Radboud University Medical Center: Molecular Neurophysiology
In our research group we aim to unravel the molecular mechanisms underlying neuronal communication at the synapse and neuronal network level. We do this through innovative and creative research in a collaborative and challenging environment by studying animal and human cellular models of neurodevelopmental disorders.
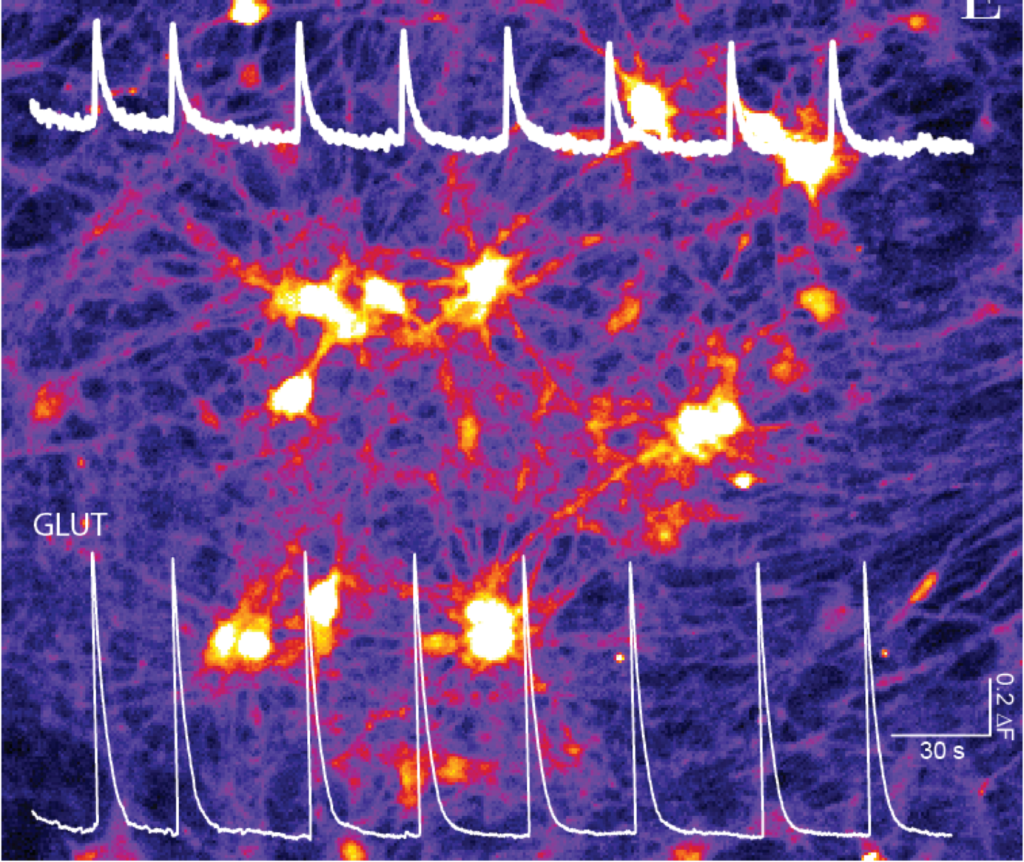
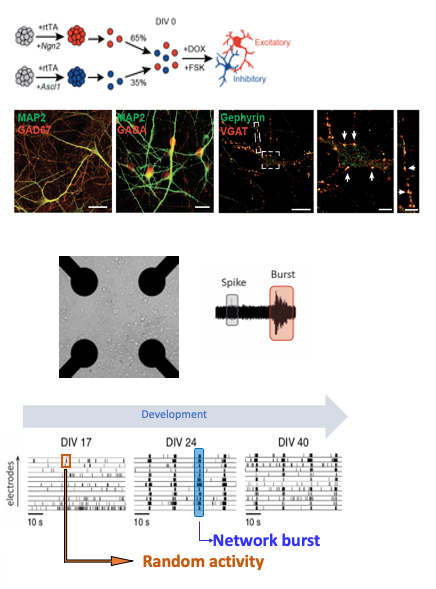
Human induced pluripotent stem cells (hIPSC) represent a novel exciting avenue to study the underlying mechanisms of neurological diseases. hIPSCs are a type of pluripotent stem cells derived from reprogramming somatic cells such as fibroblasts, and can be differentiated into different lineages, including neurons. To expedite the generation of functional neurons we implemented transcription factor-based direct conversion of iPSCs into cortical upper layer glutamatergic (excitatory, iGLU) and GABAergic (inhibitory, iGABA) neurons via forced expression of NGN2 or ASCL1, respectively (Mossink et al., 2021 Mol Psych). iGLU and iGABA neurons are differentiated independently but grown together on 24-well micro-electrode arrays at a given ratio (eg 75/25) to generate composite networks.
Upon differentiation, neurons derived from healthy controls form functional, synaptically-connected neuronal networks, exhibiting patterns of synchronized and rhythmic activity. The advantage of using MEAs is that we can repeatedly and passively record the same cultures as they traverse distinct neurodevelopmental transitions without disturbing their development. In addition, since changes in neuronal morphology, intrinsic properties or synaptic connectivity are reflected at the network level, MEA recordings provide a sensitive and unbiased assessment of the neuronal properties. Our lab has benchmarked the robustness and sensitivity of MEA-derived neuronal activity patterns, with patterns derived from multiple different healthy individuals functionally clustering together, while different NDD sharply segregated from each other with distinguishable fingerprint patterns that warrant the use of MEA datasets as functionally relevant readout (Frega et al., 2019 Nat comms; Klein Gunnewiek et al., 2020 Cell Rep; Mossink et al., 2021 Mol Psych; Linda et al., 2021 Autophagy). We demonstrate that iGABA neurons exert scalable inhibitory control onto glutamatergic neurons at the network level, and we developed a multiparametric analysis pipeline to characterize E/I co-culture MEA data. Using this, we defined the electrophysiological network parameters that are most strongly affected by inhibition in human in vitro neuronal networks. In BRAINMODEL we will leverage this approach to study several neurodevelopmental disorders.

VU University: The linkenkaer lab
The Linkenkaer lab has developed the critical oscillations computational model of neuronal networks to mechanistically understand how E/I regulation at the level of synaptic transmission or circuit connectivity relate to the dynamics of population activity. Thus, this computational model will facilitate predictions of how cellular phenotypes identified in vitro translate into EEG phenotypes in children with neurodevelopmental disorders and, possibly, how pharmacological intervention can restore balanced population dynamics.
University of Twente: The Clinical Neurophysiology group
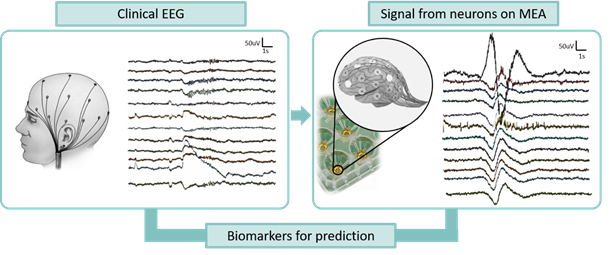
The Clinical Neurophysiology group at the UT is composed by biologists, engineers, and clinical doctors with daily, close collaboration. Our multi-disciplinary group performs translational research for neurological disorders.
The group has strong expertise in in vitro neuronal models (of rodent and human origin), Micro-electrode array (MEA) technology, computational models, EEG signal recording and analysis. The group will contribute to the consortium by integrating the data obtained from the multi-level strategy with the aim of finding biomarkers for treatment prediction.

Amsterdam UMC: N=You center, Emma Children’s Hospital
The ultimate goal of BRAINMODEL is to inform new, rational treatments for children with severe forms of neurodevelopmental disorders. Some of these treatments will be based upon existing drugs (repurposing) and immediately clinically applicable. N=You serves as the clinical hub where BRAINMODEL will apply these newly indicated treatments in children. The center is based in the Emma Children’s Hospital at AUMC and is the nation-wide referral for precision interventions in neurodevelopmental disorders.
PRECISION MEDICINE entails a paradigm shift from a one-size-fits-all to a tailored and personalized approach in medicine. This is relevant for early diagnosis and to deliver the right treatment in the right way at the right moment to enable a better prognosis for patients. It holds the promise of improving quality of life and, simultaneously achieving a reduction of health care costs.
Following extensive preclinical and clinical studies, we have optimized our N=You precision method into three stages of treatment:
- Informing – linking the right patient to the right drug by integration of cellular, genetic, EEG and clinical information in ‘N=You’ profiles.
- Performing – applying the personalized treatment in the appropriate n-of-1 design where the child serves as his or her own control
- Monitoring – following up treatment success by repeated measurement of EEG, cognitive and patient-report outcome measures
Patients and caregivers can follow the course of the N=You trajectory on an individualized dashboard and report PROMs via the Klik user friendly portal. The outcome and success of these treatments will be communicated back to BRAINMODEL researchers for further optimization of the cellular and neurophysiological diagnostic tests.
Hilgo Bruining is the clinical director of the Center and associate professor in child and adolescent psychiatry.

Radboud UMC: The center of expertise in rare genetic neurodevelopmental disorders; Department of Human Genetics & Vincent van Gogh institute for Psychiatry: Centre of Excellence for Neuropsychiatry
Our key ambition is to improve diagnostics, management and treatment of patients with monogenetic causes of neurodevelopmental disorders, also referred to as rare Mendelian syndromes. Although these disorders likely affect 1-3% of the population and disrupt daily lives of patients and their caregivers, therapeutic management presently lacks effective intervention tools which is mainly due to the rareness of individual mutations. However, next generation sequencing has revolutionized identification of causative genetic mutations in many Mendelian syndromes. For a considerable number of individuals, a molecular diagnosis can increasingly be provided. As a consequence, it is now possible to identify individuals sharing the same molecular defect. The children diagnosed with these syndromes are so far largely understudied and do not benefit from specific, personalized treatment. Therefore findings derived from pathological mechanisms brought about by these individual and specific gene mutations now urgently need translation to clinical care. Within BRAIN MODEL Tjitske Kleefstra and team will fill this gap and have introduced to the Radboudumc center of expertise in rare genetic neurodevelopmental disorders the innovative concept of personalized healthcare for genetic syndromes. We will integrate the studies from molecular and cellular mechanisms for which we study patient derived cells (from a blood or skin sample), with clinical characterization including innovative EEG measures, to obtain individual treatment tools. Our multidisciplinary team will carefully collaborate with neuroscientists from the RUMC and AUMC and with researchers and clinicians from the N=You clinic in AUMC to perform the cellular studies and clinical characterization and treatment in a joint effort.

Radboud University: The department of Synthetic Organic Chemistry
Floris Rutjes is Head of the Department of Synthetic Organic Chemistry within the Institute for Molecules and Materials at Radboud University. The group has ample expertise in the synthesis of small organic molecules with a focus on biological activity. This expertise is exploited in three different ways:
- Development of new synthesis methodology and/or techniques that can be used for synthesizing new structural moieties and/or conjugation methods.
- Development of (tracer) molecules that can aid in elucidating and potentially interfere with biological mechanisms at the molecular level.
- Development of novel lead compounds to modulate (activate or inhibit) biological targets that are relevant for specific diseases: this concerns the optimization of (often heteroaromatic) small molecules in iterative cycles of (computer-aided) design, synthesis and biological testing to reach suitable in vitro and (preclinical) in vivo properties (hit-to-lead development, lead optimization).
The latter expertise will also be our contribution to BRAINMODEL with the aim to identify and optimize small organic molecules that can modulate the cellular targets that are studied in this program.
University of Twente: The Mesoscale Chemical Systems group and AMBER group
The Mesoscale Chemical Systems group at the UT, together with the AMBER group and in collaboration with the Synaptic Computation group at the Amsterdam UMC – Location VUmc, will contribute by developing on-chip multi-cell patch-clamp technology. These groups will combine their expertise in micro/nanofabrication, microfluidics and electrophysiology to realize chips to perform parallel electrical read-out on cultured networks of neuronal cells. The “neurochip” will be used to speed up cellular analysis of patient derived neurons by the Verhage and Cornelisse team and for screening of novel E/I targeting drugs, developed by the Rutjes team, in these cells. Important challenges include on-chip automated cell trapping and Giga-seal formation.






